Introduction
Mutation can be described as the permanent alteration of the nucleotide sequence of the genome of an organism, virus, or extrachromosexual DNA or other genetic elements. The first step in the emergence of resistance is a genetic change in a bacterium, and there are two ways this can happen:
- Transfer of antibiotic-resistant genes: This is when an existing antibiotic-resistant gene transfers from one bacterium to another bacterium.
- Spontaneous mutation in the bacterium’s DNA: Most antibiotics work by inactivating an essential protein. Genetic change can remove this protein. Moreso, mutations in the target protein can prevent the antibiotic from binding it from inactivating the target protein.
Streptomycin is an inhibitor of bacterial protein synthesis and streptomycin resistance is frequently due to mutations in the gene encoding the ribosomal protein S12, rpsL.
Materials Needed
- Nutrient Agar 15ml per bottle
- Sterile Petri dishes
- Sterile streptomycin solution 5mg/ml 10mg/ml, 15mg/ml, 20mg/ml, 25mg/ml
- 18 – 24hrs broth culture of E-coli
Procedures
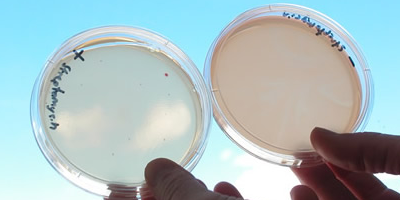
- Prepare two gradient plates as follows: One with streptomycin, and the other without. Pour one bottle of melted cooled (Molten) nutrient agar unto a Petri dish, elevate one side of the plate so that the medium is in thin layer on one side and a thick layer on the other.
- When this layer of medium solidifies, place on a horizontal surface and pour in a bottle of molten nutrient agar to which you previously added 1ml sterile streptomycin solution and to the other plate you added ordinary melted nutrient agar without streptomycin. The streptomycin will establish a concentration gradient across the plate; the high concentration of streptomycin will be where the streptomycin agar is thick and low concentration on the opposite side. Please mark your plate accordingly.
- Inoculate each plate by placing a drop of the E-coli culture on the agar surface, the spread it out uniformly using a sterile glass-spreader.
3a). Setup a control (without streptomycin) - Incubate at 37oC for 2-4 days
- Observe for development of resistant colonies in the area of higher streptomycin concentration. With a sterile loop make a suspension of cells in broth from colonies growing in the region of highest antibiotic concentration.
- Using replica plating method, transfer colonies observed in (5) to nutrient agar plates containing different concentration of the antibiotic (5mg – 25mg/ml) as stated in the Materials Needed section above. Indicate a plate with no antibiotic to serve as control.
- Incubate all plates at 37oC for at least 18 hours and observe for resistant / persister colonies.
References
- Obafemi Awolowo University Microbiology Laboratory Manual
Download “Isolation of Streptomycin-Resistant Mutant”
Isolation-of-Streptomycin-Resistant-Mutant.docx – Downloaded 0 times – 14.19 KB